SDF to Excel file, in an automated fashion
Sharing SDF files between chemists is often a pain. It’s supposed to be vanilla and super-standard but sometimes still gives everyone involved a headache. Especially when moving SDF between two chemistry codes, especially if hydrogens are involved…
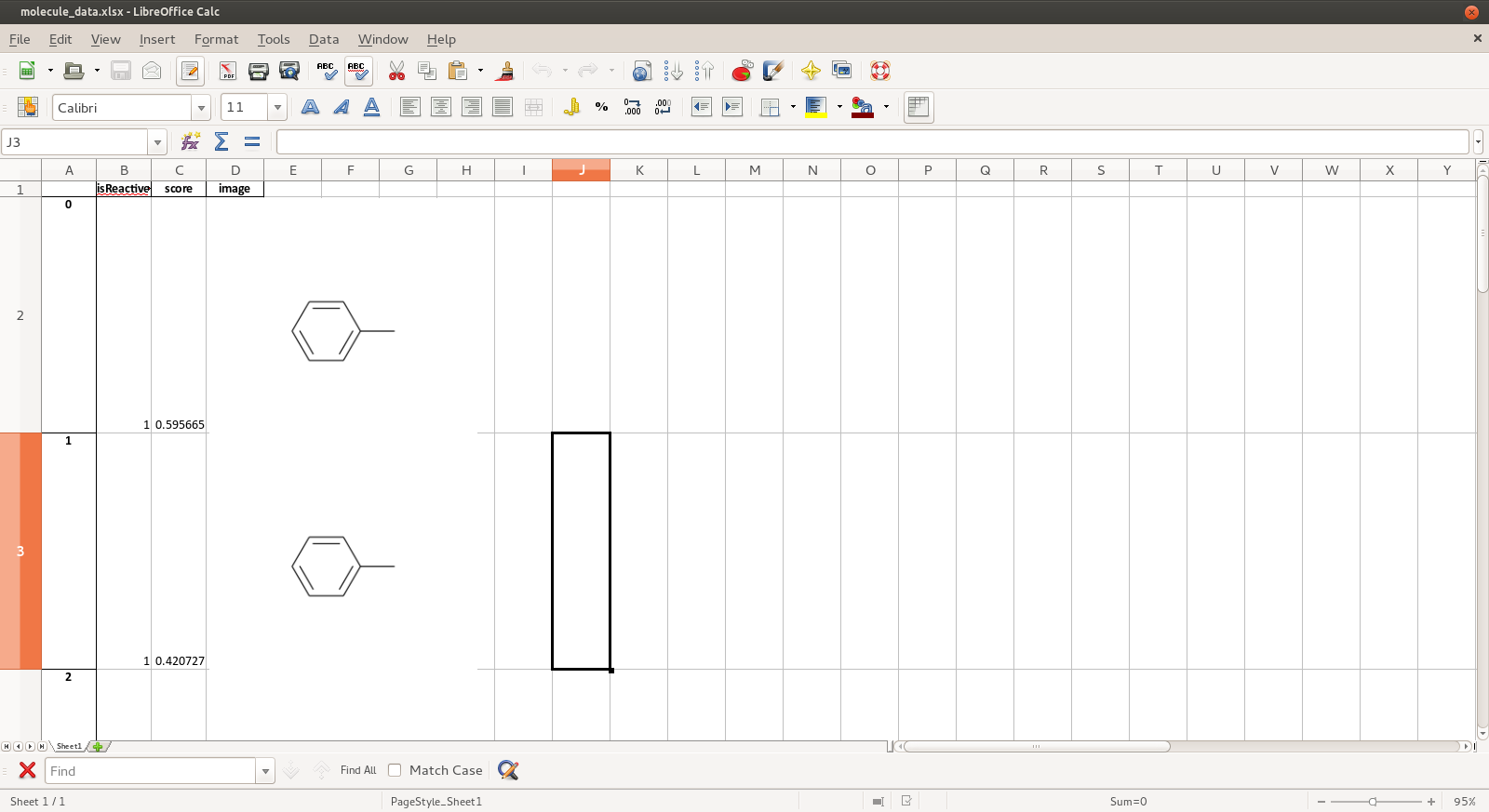
For this reason, and because some people ONLY work with excel files, it’s good to have an ability to automatically convert an SDF file to a Excel file (especially xlsx). With pandas and rdkit, its possible to easily make such moves. Example below.
from rdkit import Chem
from rdkit.Chem import Draw
import pandas as pd
import random
from random import randrange
# Create some fake molecules
ms = [Chem.MolFromSmiles('Cc1ccccc1') for i in range(10)]
# Set a fake 'score' property
for m in ms:
m.SetProp("score", str(randrange(1)))
# Set a fake 'isReactive' flag
for m in ms:
m.SetProp("isReactive", str(bool(random.getrandbits(1))))
# Convert to a pandas DataFrame
# (i know this is a little contrived but that's
# the best one can do without a 'real' sdf)
def to_df(ms):
props = ms[0].GetPropNames()
data = [[m] + [m.GetProp(p) for p in props] for m in ms]
df = pd.DataFrame(data, columns=["mol", "isReactive", "score"])
df.isReactive = map(bool, df.isReactive)
df.score = map(float, df.score)
return df
df = to_df(ms)
# Create image files to be embedded,
# JD: this is clumsy, we should be passing
# Image objects rather than filenames...
image = []
for i, mol in enumerate(df["mol"]):
f = "molecule_{}.png".format(i)
Draw.MolToFile(mol, f)
image.append(f)
df['image'] = image
# Remove molecules column, rdkit.Mol objects cannot be saved to excel :(
del df['mol']
writer = pd.ExcelWriter('molecule_data.xlsx', engine='xlsxwriter')
df.to_excel(writer, sheet_name='Sheet1')
writer.close()
Pandas uses xlsxwriter module to support the Excel format. There is no easy way to pass image objects, embedded in the pandasa.DataFrame, down to xlsxwriter. The writer itself supports the insert_image functionality that takes a filename as argument example). The easiest way is to make pandas detect that a cell contains a string ending with a .png and take use ‘insert_image’, see the hack below:
# directly editng ~/.local/lib/python2.7/site-packages/pandas-0.13.1-py2.7-linux-x86_64.egg/pandas/io/excel.py
if cell.mergestart is not None and cell.mergeend is not None:
wks.merge_range(startrow + cell.row,
startcol + cell.col,
startrow + cell.mergestart,
startcol + cell.mergeend,
cell.val, style)
# this is the new hack
elif type(cell.val) is str and cell.val.endswith(".png"):
wks.insert_image(startrow + cell.row, startcol + cell.col, cell.val)
wks.set_row(cell.row, 200)
wks.set_column(cell.col, 400)
else:
wks.write(startrow + cell.row,
startcol + cell.col,
cell.val, style)
And here you go: molecule_data.xlsx has a beautiful column with molecule images. There is one catch: one needs to modify pandas a tiny-tiny bit…